Applications
Analysis of various low-complexity proteins using SubSeqer
Elastin
Previously we have successfully applied the methodology behind Subseqer to elastin, a well-studied low-complexity extracellular protein which imparts elastomeric properties to various tissues including arteries, lung and skin (Mithieux et al 2005). The monomeric unit of elastin is composed of alternating cross-linking and hydrophobic domains (Li et al 2002). Using MEME, we were able to successfully identify the more rigidly defined repetitive elements of the cross-linking domain. However, the less well defined motifs which are thought to play a vital role in the allowing the hydrophobic regions to impart elasticity to the protein were not found. By using methods outlined in this paper on a number of elastin sequences from various species, we discovered several over-represented motifs such as G*VPG in all elastins, and GGLGTGGLP in Zebrafish elastin (He et al 2007).
Resilin
Resilin is an elastic protein found in insect cuticles, providing energy storage and vibration dampening (Gosline et al 2002). Using the 575 amino acid resilin protein from Drosophila melanogaster (Accession: NP_995860.1) and SubSeqer, we found two repetitive domains interspersed by a non repetitive region. The first repetitive domain was composed of repeats of a GGRPS[SS|DT]VGAPG*G*G motif, and the second repetitive domain was composed of GYS[G|S]GRPGG repeats. This motif confirms previous characterization of the resilin protein (Tatham, el al 2002) and showcases the effectiveness of the Subseqer software.
Flagelliform Spider Silk
Flagelliform spider silk forms the spirals of spider webs that are used to capture prey (Hayashi et al 2001). Unlike the dragline spider silks which have great tensile strength, flagelliform silks are more elastic1. For a 907 amino acid flagelliform protein from Nephila clavipes (Accession: AAC38847.1) SubSeqer found that the protein is made up of long stretches of GPGGS, GPGGY, GPGGA, and GPGGV repeats.
Wheat Gluten
Unlike resilin, elastin, and spider silk, which have evolved to provide elasticity, gluten proteins, whose primary role is energy storage in germinating cells, appear to have elastomeric properties strictly as a result of their sequence (Shewry 2002). Using the 661 amino acid HMW subunit from Aegilops searsii (Accession: AAT38813.1) we found this protein was composed primarily of a 14 amino acid motif QQPGQGQQG[YH]YP[TA]S.
The observation that the PG dipeptide is a frequently a hub in all of the proteins studied suggests that this particular pair of amino acids in this exact order is necessary for providing structural proteins with their unique characteristics. Using SubSeqer it is possible to identify other such hubs when studying and comparing proteins from multiple species or multiple proteins from a single species. SubSeqer provides the flexibility to examine repetitive sequences through informative visual inspection, a perspective which till now has been unavailable.
Analysis of elastin sequences using other algorithms
Existing tools such as Teiresias and MEME are motif finding algorithms aimed at identifying consensus motifs within families of proteins that can typically be used to help classify them and/or identify new examples of proteins belonging to those families. On the other hand, SubSeqer is concerned at identifying structurally and/or functionally relevant motifs within individual sequences (i.e. it does not compare across families). While it is possible to apply tools like Teiresias/MEME to families of these low complexity proteins, due to the inherent variability associated with these sequences, identification of biologically relevant motifs is problematic. For many such proteins it is the ‘style’ of sequence rather than the sequence composition which is conserved. By adopting a subsequence approach, SubSeqer is better able to identify small motifs of functional/structural relevance from within a variety of different sequence contexts that arise as a consequence of low sequence complexity. Furthermore, the integration of the WebLogo tool also enables SubSeqer to provide additional sequence context compared with uninformative wild cards.
Teiresias
Teiresias uses heuristics to consider all possible motifs in a bottom-up approach and finds long, simple patterns using an assembly of shorter patterns. Applying Teiresias to 15 previously identified elastin sequences resulted in the following top five motifs.The consensus column shows the detected pattern with the '.' character representing wildcards and the P-values for each pattern are shown in the second column.
| Consensus | P-value |
| PGVPLGYPIK.PKLPG | 1.28e-94 |
| G.GVLPGV.TG.G.K.K.PG.G | 1.66e-84 |
| F.G.GVLPGV.TG.G.K.K | 1.20e-96 |
| PG..PG.G.PG..PGG..PG | 3.93e-71 |
| G.GVLPGV.TG.G.K.K | 1.60e-88 |
Of these three are essentially the same (G.GVLPGV.TG.G.K.K). From inspection of these motifs, it is not clear what the key structural/functional units are. From the 4th motif, one may deduce that the PG dipeptide may play an important role in the structure of the protein, however the wildcard characters do not provide sufficient context about this dipeptide. The other motifs are longer than those suggested by the elastin literature to form the major structural/functional units. In addition, Teiresias failed to detect the obvious and conserved poly-alanine regions commonly found in the cross-linking regions of elastin.
MEME
MEME detects gapless motifs using position-dependent letter-probability matrices which describe the probability of each possible letter at each position in the pattern. When MEME is applied to the same 15 elastin sequences, it returns the following five ‘best’ motifs (see table below). With the exception of the third motif (VLPGV.TG – also found by Teiresias) it is difficult to interpret what the key structural/functional units are. The third motif is particularly misleading since it is known that many elastin sequences contain many repeats of a KAAK / KAAAK known to be involved in forming cross-links between adjoining proteins.
| Consensus | P-value |
| .G.FG..QPGVPLGYPIKAPKLP | 1.78e-179 |
| ..GK.CCR.RK | 3.75e-142 |
| .......VLPGV.TG...... | 1.22e-124 |
| ..................K.G.. | 3.43e-127 |
| ..F..GGVA.RPGFGLSPI.PGG | 1.09e-90 |
SubSeqer
Unlike Teiresias and MEME, SubSeqer does not attempt to identify motifs through the analysis of families of protein sequences, but rather attempts to identify key structural and/or functional repetitive elements within individual sequences. One of the key concepts underlying SubSeqer is the identification of motifs through the combination of smaller subsequences. Particularly for low complexity proteins, many different motifs may share similar sequences that provide distinct structural and/or functional roles.
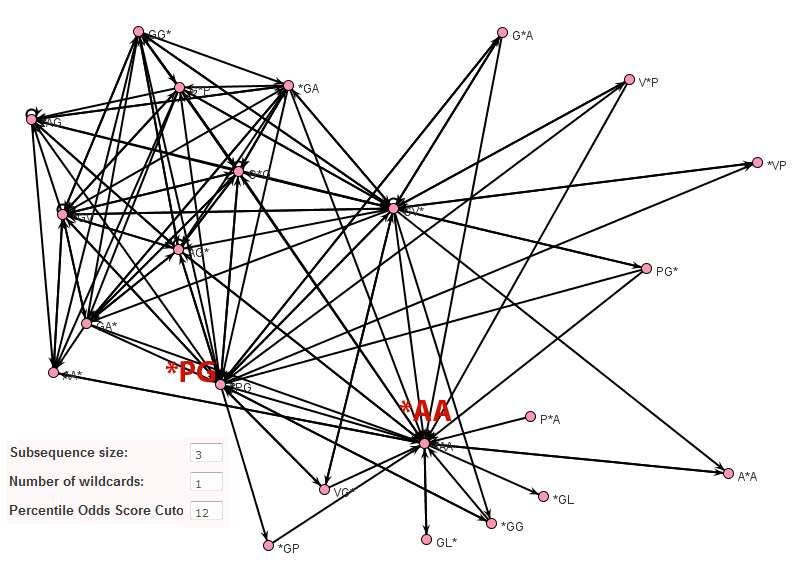
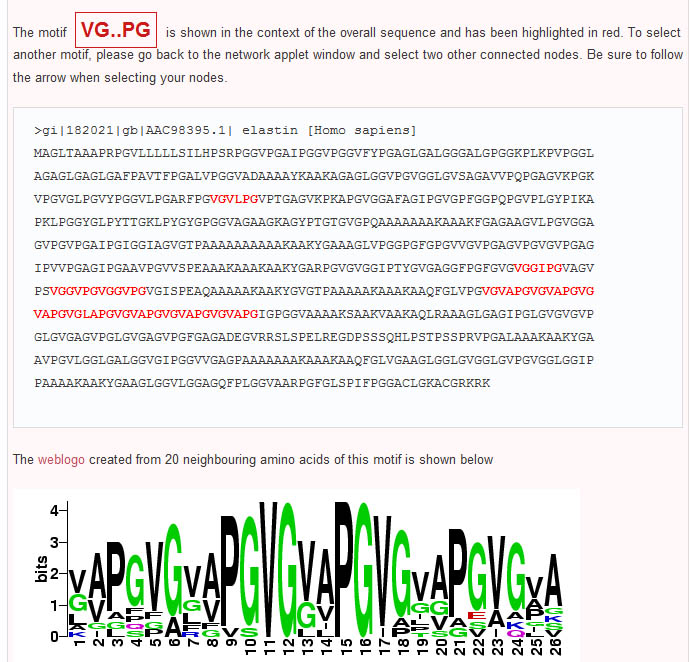
In contrast to Teiresias and MEME, the subsequence adjacency network representation of elastin produced by Subseqer (shown below), clearly identifies "hub" subsequences such as *PG and *AA that are integral to a number of possible motifs. Previous studies have noted that the dipeptide - PG is associated with and is likely required for the initiation of beta-turns (Urry and Parker, J. Muscl. Res. Cell Motil. 2002). Further expansion of these motifs using sequence logos provides the context of these motifs. For instance, when the nodes VG* and PG* are chosen to build a sequence logo, it is clear that this VP*PG pentamer motif is really part of a repetitive [GV][AV]PGVG motif.