PhyloPro - Applications
One of the major uses of PhyloPro is visualizing the evolutionary trajectories of components
of biological systems such as protein complexes, signaling or metabolic pathways. Indeed,
applying PhyloPro to any protein set, however derived, may provide potentially useful
conservation information. Applying PhyloPro, the user is readily able to identify components
that are well conserved as well as those that may represent taxon-specific innovations. The
ability to subsequently download putative orthologs may also facilitate more in depth
phylogenetic analyses. Previously we have successfully applied PhyloPro to investigate
patterns of conservation in chromatin remodelling complexes [On et al., 2010], ubiquitin
ligase substrates [Persaud et al., 2009] and the extracellular matrix [Cromar et al., in
preparation]. Below we illustrate an example where PhyloPro has been used to examine the
conservation of 29 proteins associated with glycerophospholipid metabolism biosynthesis in
yeast. These proteins were obtained from simply querying the KEGG database resource.
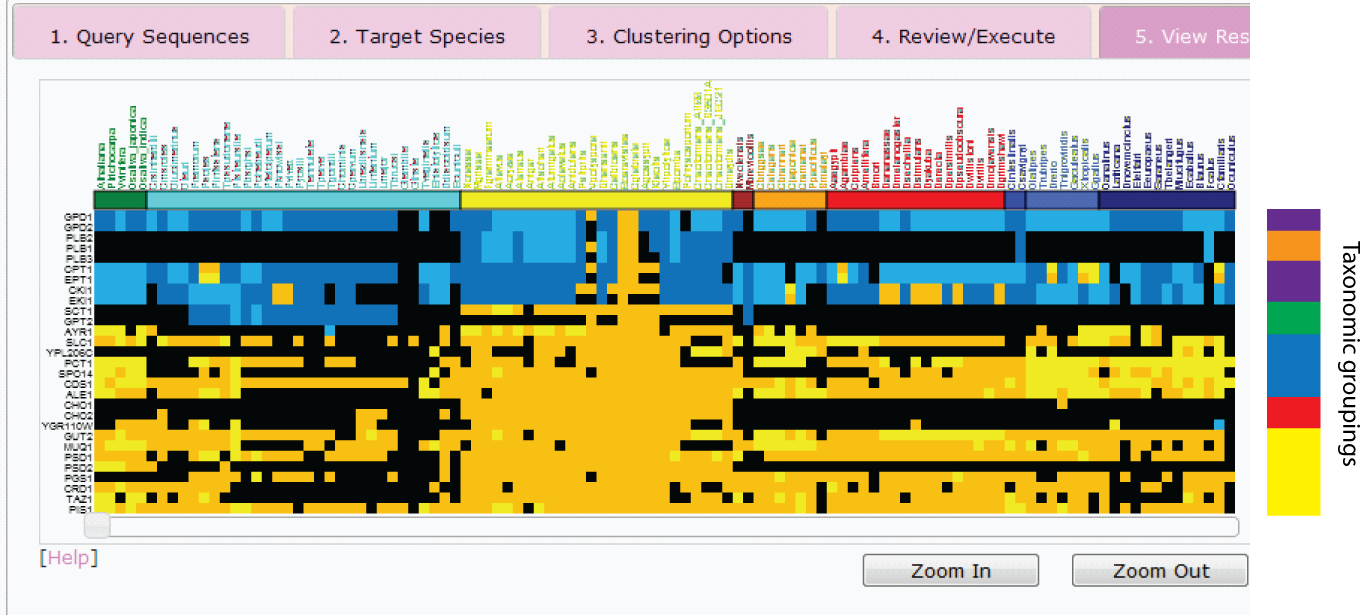
From the above heatmap it is possible to identify several clusters of proteins with similar
patterns of conservation. For example, PLB1,2 and 3 (orange cluster) represent sequences
that appear specific to fungi. Note that there may indeed be orthologs in other species,
however such orthologs are not detectable through our InParanoid pipeline and hence one may
deduce that the sequences at the very least have undergone significant divergence in the
fungal lineage. On the other hand, proteins in the blue cluster (e.g. PCT1, SPO14 and COS1)
appear to have undergone significant expansion within the deuterstomes (species coloured in
blue at the top right of the heatmap). Using the colourmap tool available as part of the
KEGG database resource, we can map these
cluster patterns onto the metabolic map as shown below.
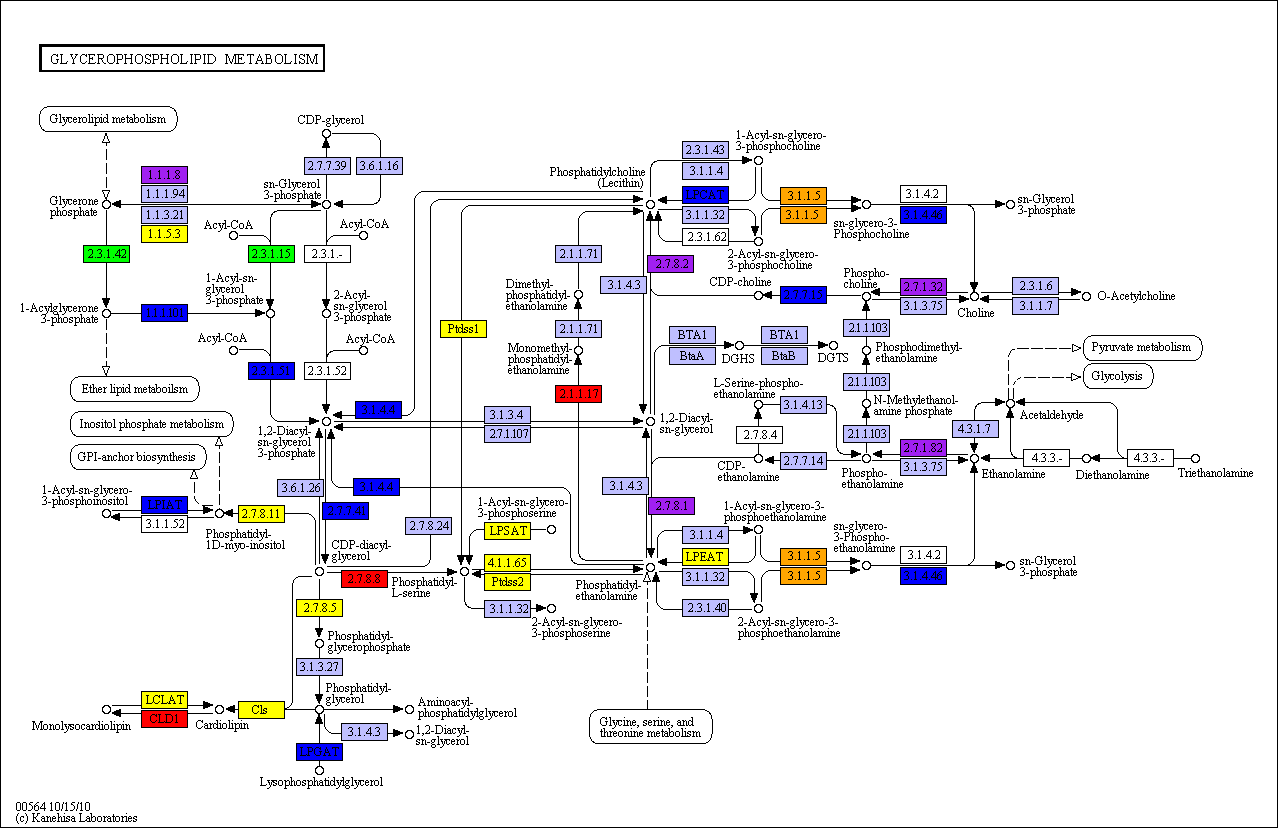
From the above, we see that the highly conserved proteins (yellow, purple and blue enzymes)
are associated with maintaining a core set of reactions within the pathway linking to
phosphatidyl ethanolamine, phosphatidyl choline, phosphatidyl inositol and phosphatidyl
serine. In a similar vein, we mapped conservation profiles of chromatin modification
proteins onto their complexes to reveal highly conserved catalytic cores associated with
taxon-specific regulatory elements [On et al., 2010].
References:
Persaud, A., Amsen, E.M.,Xiong, X., Wasmuth, J.,Saadon, Z., Fladd, C., Parkinson, J., and
Rotin, D. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and
Nedd4-2 using proteome arrays. Molecular Systems Biology. 5:333
On, T., Xiong, X., Pu, S., Turinsky, A., Gong, Y., Emili, A., Zhang, Z., Greenblatt, J.,
Wodak, SJ. and Parkinson, J. (2010) The evolutionary landscape of the chromatin modification
machinery. Proteins. 78:2075-2089
Cromar, C.L. and Parkinson, J. Surveying the Extracellular Matrix: Towards a systems level
understanding of its structure, function and evolution. In preparation.
Wan, C., Borgeson, B., Phanse, S., Tu, F., Drew, K., Clark, G., Xiong, X., Kagan, O., Kwan, J.,
Berzginov, A., Chessman, K., Pal, S., Cromar, G., Papoulas, O., Ni, Z., Boutz, DR.,
Snejana Stoilova, S., Havugimana, P., Guo, X., Malty, R., Sarov, M., Greenblatt, J., Babu, M.,
Derry, WB., Tillier, E., Wallingford, J., Parkinson, J., Marcotte, E. and Emili, A. (2015).
Panorama of ancient metazoan macromolecular complexes. Nature. 525(7569):339-44.

